References

Abecassis, M., Senina, I., Lehodey, P., Gaspar, P., Parker, D., Balazs, G., Polovina, J. (2013). A Model of Loggerhead Sea Turtle (Caretta caretta) Habitat and Movement in the Oceanic North Pacific. PLoS ONE, 8(9), e73274, https://doi.org/10.1371/journal.pone.0073274
Allen, J. I., M. Eknes, and G. Evensen. (2003). An Ensemble Kalman Filter with a complex marine ecosystem model: hindcasting phytoplankton in the Cretan Sea. Annales Geophysicae, 21(1), 399-411, https://doi.org/10.5194/angeo-21-399-2003
Almroth-Rosell, E., Eilola, K., Kuznetsov, Hall, I.P.O.J., and Meier, H.E.M. (2015). A new approach to model oxygen dependent benthic phosphate fluxes in the Baltic Sea. Journal of Marine Systems, 144, 127-141, https://doi.org/10.1016/j.jmarsys.2014.11.007
Anderson, L.A., Robinson, A.R., Lozano, C.J. (2000). Physical and biological modeling in the Gulf Stream region: I. Data assimilation methodology. Deep Sea Research Part I: Oceanographic Research Papers, 47, 1787-1827, https://doi.org/10.1016/S0967-0637(00)00019-4
Audzijonyte, A., Pethybridge, H., Porobic, J., Gorton, R., Kaplan, I., Fulton, E.A. (2019). Atlantis: A spatially explicit end-to-end marine ecosystem model with dynamically integrated physics, ecology and socio-economic modules. Methods in Ecology and Evolution, 10, 1814-1819, https://doi.org/10.1111/2041-210X.13272
Aumont, O., Ethé, C., Tagliabue, A., Bopp, L., and Gehlen, M. (2015). PISCES-v2: an ocean biogeochemical model for carbon and ecosystem studies. Geoscientific Model Development, 8, 2465-2513, https://doi.org/10.5194/gmd-8-2465-2015
Aumont O., Maury O., Lefort S., Bopp L. (2018). Evaluating the Potential Impacts of the Diurnal Vertical Migration by Marine Organisms on Marine Biogeochemistry. Global Biogeochemical cycles, 32 (11), 1622-1643, https://doi.org/10.1029/2018GB005886
Baretta, J. W., Ebenhöh, W., and Ruardij, P. (1995). The European regional seas ecosystem model, a complex marine ecosystem model. Netherlands Journal of Sea Research, 33, 233-246, https://doi.org/10.1016/0077-7579(95)90047-0
Bell, J.D., Ganachaud, A., Gehrke, P.C., Griffiths, S.P., Hobday, A.J., Hoegh-Guldberg, O., Johnson, J.E., Le Borgne, R., Lehodey, P., Lough, J.M., Matear, R.J., Pickering, T.D., Pratchett, M.S., Sen Gupta, A., Senina, I. and Waycott, M. (2013). Mixed response of tropical Pacific fisheries and aquaculture will respond differently to climate change. Nature Climate Change, 3, 591-599, https://doi.org/10.1038/nclimate1838
Bell, J.D., Senina, I., Adams, T., Aumont, O., Calmettes, B., Clark, S., Dessert, M., Hampton, J., Hanich, Q., Harden-Davies, H., Gehlen, M., Gorgues, T., Holmes, G., Lehodey, P., Lengaigne, M., Mansfield, B., Menkes, C., Nicol, S., Pasisi, C., Pilling, G., Ota, Y., Reid, C., Ronneberg, E., Sen Gupta, A., Seto, K., Smith, N., Taei, S., Tsamenyi, M., Williams, P. (2021). Pathways to sustaining tuna-dependent Pacific Island economies during climate change. Nature Sustainability, 4, 900-910, https://doi.org/10.1038/s41893-021-00745-z
Berline, L., Brankart, J.M., Brasseur, P., Ourmières, Y., and Verron, J. (2007). Improving the physics of a coupled physical–biogeochemical model of the North Atlantic through data assimilation: Impact on the ecosystem. Journal of Marine Systems, 64(1-4), 153-172, https://doi.org/10.1016/j.jmarsys.2006.03.007
Berthet, S., Séférian, R., Bricaud, C.,Chevallier, M., Voldoire, A., and Ethé, C. (2019). Evaluation of an online grid-coarsening algorithm in a global eddy-admitting ocean biogeochemical model. Journal of Advances in Modeling Earth Systems, 11, 1759-1783, https://doi.org/10.1029/2019MS001644
Bertino, L., Evensen, G., and Wackernagel, H. (2003). Sequential data assimilation techniques in oceanography. International Statistical Review, 71(2), 223-241, http://www.jstor.org/stable/1403885.
Biogeochemical-Argo Planning Group (2016). The Scientific Rationale, Design andImplementation Plan for a Biogeochemical-Argo Float Array. https://doi.org/10.13155/46601
Blanchard, J.L., Heneghan, R.F., Everett, J.D., Trebilco, R., Richardson, A.J. (2017). From bacteria to whales: Using function size spectra to model marine ecosystems. Trends in Ecology and Evolution, 32(3), 174-186, https://doi.org/10.1016/j.tree.2016.12.003
Boyer, T.P., Baranova, O.K., Coleman, C., Garcia, H.E., Grodsky, A., Locarnini, R.A., Mishonov, A.V., Paver, C.R., Reagan, J.R., Seidov, D., Smolyar, I.V., Weathers, K., Zweng, M.M. (2018). World Ocean Database 2018. A.V. Mishonov, Technical Ed., NOAA Atlas NESDIS 87, https://www.ncei.noaa.gov/products/world-ocean-database
Brankart, J.-M., Testut, C.-E., Béal, D., Doron, M., Fontana, C., Meinvielle, M., and Brasseur, P. (2012). Towards an improved description of oceanographic uncertainties : effect of local anamorphic transformations on spatial correlations. Ocean Science, 8, 121-142, https://doi.org/10.5194/os-8-121-2012
Brasseur, P., Gruber, N., Barciela, R., Brander, K., Doron, M., El Moussaoui, A., Hobday, M. Huret, A.J., Kremeur, A.-S., Lehodey, P., and others. (2009). Integrating biogeochemistry and ecology into ocean data assimilation systems. Oceanography, 22(3), 206-215, https://doi.org/10.5670/oceanog.2009.80
Bricaud, C., Le Sommer, J., Madec, G., Calone, C., Deshayes, J., Éthé, C., Chanut, J., Lévy, M. (2020). Multigrid algorithm for passive tracer transport in the NEMO ocean circulation model: a case study with the NEMO OGCM (version 3.6). Geoscientific Model Development, 13(11), 5465-5483, https://doi.org/10.5194/gmd-13-5465-2020
Brown, C. D., and Davis, H. T. (2006). Receiver operating characteristics curves and related decision measures: A tutorial. Chemometrics and Intelligent Laboratory Systems, 80(1), 24-38 https://doi.org/10.1016/j.chemolab.2005.05.004
Butenschön, M., Clark, J., Aldridge, J. N., Allen, J. I., Artioli, Y., Blackford, J., Bruggeman, J., Cazenave, P., Ciavatta, S., Kay, S., Lessin, G., van Leeuwen, S., van der Molen, J., de Mora, L., Polimene, L., Sailley, S., Stephens, N., and Torres, R. (2016). ERSEM 15.06: a generic model for marine biogeochemistry and the ecosystem dynamics of the lower trophic levels. Geoscientific Model Development, 9, 1293-1339, https://doi.org/10.5194/gmd-9-1293-2016
Campbell, J. (1995). The lognormal distribution as a model for bio-optical variability in the sea. Journal of Geophysical Research: Oceans, 100, C7, 13237-13254, https://doi.org/10.1029/95JC00458
Capet, A., Meysman, F. J. R., Akoumianaki, I., Soetaert, K., and Grégoire, M. (2016). Integrating sediment biogeochemistry into 3-D oceanic models: A study of benthic-pelagic coupling in the Black Sea. Ocean Modelling, 101, 83-100, https://doi.org/10.1016/j.ocemod.2016.03.006
Carlotti, F., Wolf, K.U. (1998). A Lagrangian ensemble model of Calanus finmarchicus coupled with a 1-D ecosystem model. Fisheries Oceanography, 7,191-204, https://doi.org/10.1046/j.1365-2419.1998.00085.x
Carrassi, A., Bocquet, M., Bertino, L., and Evensen, G. (2018). Data assimilation in the geosciences: An overview of methods, issues, and perspectives. Wiley Interdisciplinary Reviews: Climate Change, e535, https://doi.org/10.1002/wcc.535
Carroll, D., Menemenlis, D., Adkins, J.F., Bowman, K.W., Brix, H., Dutkiewicz, S., Fenty, I., Gierach, M.M., Hill, C., Jahn, O., and Landschützer, P. (2020). The ECCO-Darwin data-assimilative global ocean biogeochemistry model: Estimates of seasonal to multidecadal surface ocean pCO2 and air-sea CO2 flux. Journal of Advances in Modeling Earth Systems, 7(3-4), 191-204, https://doi.org/10.1029/2019MS001888
Chai, F., Johnson, K., Claustre, H., Xing, X., Wang, Y., Boss, E., Riser, S., Fennel, K., Schofield, O., Sutton, A. (2020). Monitoring ocean biogeochemistry with autonomous platforms. Nature Reviews Earth & Environment, 1, 315-326, https://doi.org/10.1038/s43017-020-0053-y
Chambault P., de Thoisy B., Heerah K., Conchon A., Barrioz S., Dos Reis V., Berzins R., Kelle L., Picard B., Roquet F., Le Maho Y., Chevallier D. (2016). The influence of oceanographic features on the foraging behavior of the olive ridley sea turtle Lepidochelys olivacea along the Guiana coast. Progress in Oceanography, 142: 58-71, https://doi.org/10.1016/j.pocean.2016.01.006
Christensen, V., and Walters, C. (2004). Ecopath With Ecosim: Methods, Capabilities and Limitations. Ecological Modelling, 172, 109-139, https://doi.org/10.1016/j.ecolmodel.2003.09.003
Ciavatta, S., Torres, R., Saux-Picart, S. and Allen, J.I. (2011). Can ocean color assimilation improve biogeochemical hindcasts in shelf seas? Journal of Geophysical Research: Oceans, 116(C12), https://doi.org/10.1029/2011JC007219
Ciavatta, S., Torres, R., Martinez-Vicente, V., Smyth, T., Dall’Olmo, G., Polimene, L., and Allen, J. I. (2014). Assimilation of remotely-sensed optical properties to improve marine biogeochemistry modelling. Progress in Oceanography, 127, 74-95, https://doi.org/10.1016/j.pocean.2014.06.002
Ciavatta, S., Kay, S., Saux-Picart, S., Butenschön, M. and Allen, J.I. (2016). Decadal reanalysis of biogeochemical indicators and fluxes in the North West European shelf-sea ecosystem. Journal of Geophysical Research: Oceans, 121(3), 1824-1845, https://doi.org/10.1002/2015JC011496
Ciavatta, S., Brewin, R. J. W., Skákala, J., Polimene, L., de Mora, L., Artioli, Y., and Allen, J. I. (2018). Assimilation of ocean-color plankton functional types to improve marine ecosystem simulations. Journal of Geophysical Research: Oceans, 123, 834-854, https://doi.org/10.1002/2017JC013490
Ciavatta, S., Kay, S., Brewin, R.J., Cox, R., Di Cicco, A., Nencioli, F., Polimene, L., Sammartino, M., Santoleri, R., Skakala, J. and Tsapakis, M. (2019). Ecoregions in the Mediterranean Sea through the reanalysis of phytoplankton functional types and carbon fluxes. Journal of Geophysical Research: Oceans, 124(10), 6737-6759, https://doi.org/10.1029/2019JC015128
Cossarini, G., Mariotti, L., Feudale, L., Mignot, A., Salon, S., Taillandier, V., Teruzzi, A., D'Ortenzio, F. (2019). Towards operational 3D-Var assimilation of chlorophyll Biogeochemical-Argo float data into a biogeochemical model of the Mediterranean Sea. Ocean Modelling, 133, 112-128, https://doi.org/10.1016/j.ocemod.2018.11.005
Daewel, U., and Schrum, C. (2013). Simulating long-term dynamics of the coupled North Sea and Baltic Sea ecosystem with ECOSMO II: Model description and validation. Journal of Marine Systems, 119-120, 30-49, https://doi.org/10.1016/j.jmarsys.2013.03.008
Dall'Olmo, G., Dingle, J., Polimene, L., Brewin, R.J.W., Claustre, H. (2016). Substantial energy input to the mesopelagic ecosystem from the seasonal mixed-layer pump. Nature Geoscience, 9, 820-823, https://doi.org/10.1038/ngeo2818
De Roos, A.M., Schellekens, T., Van Kooten, T., Van De Wolfshaar, K., Claessen, D., Persson, L. (2008). Simplifying a physiologically structured population model to a stage-structured biomass model. Theoretical Population Biology, 73, 47-62, https://doi.org/10.1016/j.tpb.2007.09.004
DeAngelis, D.L., Gross, L.J. (1992). Individual-based Models and Approaches in Ecology. Chapman & Hall, London.
Diaz, F., D Bǎnaru, P. Verley and Y.-J. Shin (2019). Implementation of an end-to-end model of the Gulf of Lions ecosystem (NW Mediterranean Sea). II. Investigating the effects of high trophic levels on nutrients and plankton dynamics and associated feedbacks. Ecological Modelling, 405, 51-68, https://doi.org/10.1016/j.ecolmodel.2019.05.004
Dickey, T.D. (1991). The emergence of concurrent high-resolution physical and bio-optical measurements in the upper ocean and their applications. Reviews on Geophysics, 29(3), 383- 413, https://doi.org/10.1029/91RG00578
Dornan, T., Fielding, S., Saunders, R.A., Genner, M.J. (2019). Swimbladder morphology masks Southern Ocean mesopelagic fish biomass. Proceedings of the Royal Society B Biological Science, 286(1903), http://dx.doi.org/10.1098/rspb.2019.0353
Doron, M., Brasseur, P., Brankart, J.M. (2011). Stochastic estimation of biogeochemical parameters of a 3d ocean coupled physical-biogeochemical model: twin experiments. Journal of Marine Systems, 87, 194-207, https://doi.org/10.1016/j.jmarsys.2013.02.007
Dragon A-C., Senina, Hintzen N.T., Lehodey P. (2018). Modelling South Pacific Jack Mackerel spatial population dynamics and fisheries. Fisheries Oceanography, 27(2), 97-113, https://doi.org/10.1111/fog.12234
Dueri S., Faugeras B., Maury O. (2012). Modelling the skipjack tuna dynamics in the Indian Ocean with APECOSM-E: Part 1. Model formulation. Ecological Modelling, 245, 41-54, https://doi.org/10.1016/j.ecolmodel.2012.02.008
Dueri, S., Bopp, L., Maury, O. (2014). Projecting the impacts of climate change on skipjack tuna abundance and spatial distribution. Global Change Biology, 20(3): 742-753, https://doi.org/10.1111/gcb.12460
Dupont, F. (2012). Impact of sea-ice biology on overall primary production in a biophysical model of the pan-Arctic Ocean. Journal of Geophysical Research: Oceans,117(C8), https://doi.org/10.1029/2011JC006983
Eilola, K., Meier, H.E.M., Almroth, E. (2009). On the dynamics of oxygen, phosphorus and cyanobacteria in the baltic sea: A model study. Journal of Marine Systems, 75, 163-184, https://doi.org/10.1016/j.jmarsys.2008.08.009
Escobar-Flores, P.C., O’Driscoll, R.L., Montgomery, J.C. (2018). Spatial and temporal distribution patterns of acoustic backscatter in the New Zealand sector of the Southern Ocean. Marine Ecology Progress Series, 592, 19-35, https://doi.org/10.3354/meps12489
Evensen, G. (1994). Sequential data assimilation with a nonlinear quasi-geostrophic model using Monte Carlo methods to forecast error statistics. Journal of Geophysical Research: Oceans, 99(C5), 10143-10162, https://doi.org/10.1029/94JC00572
Fennel, K., Gehlen, M., Brasseur, P., Brown, C. W., Ciavatta, S., Cossarini, G., Crise, A., Edwards, C. A., Ford, D., Friedrichs, M. A. M., Grégoire, M., Jones, E., Kim, H.-C., Lamouroux, J., Murtugudde, R., Perruche, C. and the GODAE OceanView Marine Ecosystem Analysis and Prediction Task Team (2019). Advancing marine biogeochemical and ecosystem reanalyses and forecasts as tools for monitoring and managing ecosystem health. Frontiers in Marine Science, 6, 89, https://doi.org/10.3389/fmars.2019.00089
Feudale, L., Teruzzi, A., Salon, S., Bolzon, G., Lazzari, P., Coidessa, G., Di Biagio, V., Cossarini, G. (2021). Product Quality Document For the Mediterranean Sea Production Centre, MEDSEA_ANALYSISFORECAST_BGC_006_014, https://doi.org/10.25423/cmcc/medsea_analysisforecast_bgc_006_014_medbfm3
Flynn, K. J., Stoecker, D. K., Mitra, A., Raven, J. A., Glibert, P. M. Hansen, P. J., Granéli, E., and Burkholder, J. M. (2013). Misuse of the phytoplankton-zooplankton dichotomy: the need to assign organisms as mixotrophs within plankton functional types. Journal of Plankton Research, 35, 3-11, https://doi.org/10.1093/plankt/fbs062
Follows, M. J., Dutkiewicz, S., Grant, S., and Chisholm, S. W. (2007). Emergent biogeography of microbial communities in a model ocean. Science, 315(5820), 1843-1846. https://doi.org/10.1126/science.1138544
Fontana, C., Brasseur, P., and Brankart, J. M. (2013). Toward a multivariate reanalysis of the North Atlantic Ocean biogeochemistry during 1998–2006 based on the assimilation of SeaWiFS chlorophyll data. Ocean Science, 9(1), 37-56, https://doi.org/10.5194/os-9-37-2013
Ford, D., and Barciela, R. (2017). Global marine biogeochemical reanalyses assimilating two different sets of merged ocean colour products. Remote Sensing of Environment, 203, 40-54, https://doi.org/10.1016/j.rse.2017.03.040
Ford, D., Key, S., McEwan, R., Totterdell, I. and Gehlen, M. (2018). Marine biogeochemical modelling and data assimilation for operational forecasting, reanalysis, and climate research. New Frontiers in Operational Oceanography, 625-652, https://doi.org/10.17125/gov2018.ch22
Ford, D. (2021). Assimilating synthetic Biogeochemical-Argo and ocean colour observations into a global ocean model to inform observing system design. Biogeosciences, 18(2), 509-534, https://doi.org/10.5194/bg-18-509-2021
Fulton, E.A. (2010). Approaches to end-to-end ecosystem models. Journal of Marine Systems 81, 171- 183, https://doi.org/10.1016/j.jmarsys.2009.12.012
Fulton, E., Smith, A., Johnson, C. (2003). Effect of Complexity of Marine Ecosystem Models. Marine Ecology Progress Series, 253:1-16, doi:10.3354/meps253001
Garcia, H.E., Weathers, K., Paver, C.R., Smolyar, I., Boyer, T.P., Locarnini, R.A., Zweng, M.M, Mishonov, A.V., Baranova, O.K., Seidov, D., and Reagan, J.R. (2018a). World Ocean Atlas 2018, Volume 3: Dissolved Oxygen, Apparent Oxygen Utilization, and Oxygen Saturation. A. Mishonov Technical Ed.; NOAA Atlas NESDIS 83, 38pp.
Garcia, H.E., Weathers, K., Paver, C.R., Smolyar, I., Boyer, T.P., Locarnini, R.A., Zweng, M.M, Mishonov, A.V., Baranova, O.K., Seidov, D., and Reagan, J.R. (2018b). World Ocean Atlas 2018, Volume 4: Dissolved Inorganic Nutrients (phosphate, nitrate and nitrate+nitrite, silicate). A. Mishonov Technical Ed.; NOAA Atlas NESDIS 84, 35pp.
Garnesson, P., Mangin, A., and Bretagnon M. (2021). Quality Information Document, Ocean Colour Production Centre, Satellite Observation Copernicus-GlobColour Products, https://catalogue.marine.copernicus.eu/documents/QUID/CMEMS-OC-QUID-009-030-032-033-037-081-082-083-085-086-098.pdf
Garnier, F., Brankart, J. M., Brasseur, P. and Cosme, E. (2016). Stochastic parameterizations of biogeochemical uncertainties in a 1/4° NEMO/PISCES model for probabilistic comparisons with ocean color data. Journal of Marine Systems, 155, 59-72, https://doi.org/10.1016/j.jmarsys.2015.10.012
Gehlen, M., Barciela, R., Bertino, L., Brasseur, P., Butenschön, M., Chai, F., Crise, A., Drillet, Y., Ford, D., Lavoie, D., Lehodey, P., Perruche, C., Samuelsen, A., and Simon, E. (2015). Building the capacity for forecasting marine biogeochemistry and ecosystems: recent advances and future developments. Journal of Operational Oceanography, 8:sup1, s168-s187, https://doi.org/10.1080/1755876X.2015.1022350
Geider, R.J., MacIntyre, H.L., and Kana, T.M. (1997). Dynamic model of phytoplankton growth and acclimation: responses of the balanced growth rate and the chlorophyll a: carbon ratio to light, nutrient-limitation and temperature. Marine Ecology Progress Series, 148, 187-200, https://doi.org/10.3354/meps148187
Germineaud, C., Brankart, J.M., and Brasseur, P. (2019). An ensemble-based probabilistic score approach to compare observation scenarios: an application to biogeochemical-Argo deployments. Journal of Atmospheric and Oceanic Technology, 36(12), 2307-2326, https://doi.org/10.1175/JTECH-D-19-0002.1
Gharamti, M.E., Tjiputra, J., Bethke, I., Samuelsen, A., Skjelvan, I., Bentsen, M., Bertino, L. (2017). Ensemble data assimilation for ocean biogeochemical state and parameter estimation at different sites. Ocean Modelling, 112, 65-89, https://doi.org/10.1016/j.ocemod.2017.02.006
Glibert, P., Mitra, A., Flynn, K., Hansen, P., Jeong, H., and Stoecker, D. (2019). Plants Are Not Animals and Animals Are Not Plants, Right? Wrong! Tiny Creatures in the Ocean Can Be Both at Once! Frontiers for Young Minds, 7:48, https://doi.org/10.3389/frym.2019.00048
Gradinger, R. (2009). Sea-ice algae: Major contributors to primary production and algal biomass in the Chukchi and Beaufort Seas during May/June 2002. Deep Sea Research Part II: Topical Studies in Oceanography, 56, 1201-1212, https://doi.org/10.1016/j.dsr2.2008.10.016
Green, D.B., Bestley, S., Corney, S.P., Trebilco, R., Lehodey, P., and Hindell, M.A. (2021). Modeling Antarctic krill circumpolar spawning habitat quality to identify regions with potential to support high larval production. Geophysical Research Letters, 48, e2020GL091206, https://doi.org/10.1029/2020GL091206
Green, D.B., Bestley, S., Trebilco, R., Corney, S.P., Lehodey, P., McMahon, C.R., Guinet, C., and Hindell, M.A. (2020). Modelled mid-trophic pelagic prey fields improve understanding of marine predator foraging behaviour. Ecography, https://doi.org/10.1111/ecog.04939
Gregg, W.W., and Rousseaux, C.S. (2016). Directional and spectral irradiance in ocean models: effects on simulated global phytoplankton, nutrients, and primary production. Frontiers in Marine Science, 3, 240, http://dx.doi.org/10.1088/1748-9326/ab4667
Gregg, W.W., and Rousseaux, C.S. (2019). Global ocean primary production trends in the modern ocean color satellite record (1998-2015). Environmental Research Letters,14(12),124011, https://doi:10.1088/1748-9326/ab4667
Grégoire, M., Raick, C., and Soetaert, K. (2008). Numerical modelling of the central black sea ecosystem functioning during the eutrophication phase. Progress in Oceanography, 76, 286 333, https://doi.org/10.1016/j.pocean.2008.01.002
Grégoire, M., Soetaert, K. (2010). Carbon, nitrogen, oxygen and sulfide budgets in the Black Sea: a biogeochemical model of the whole water column coupling the oxic and anoxic parts. Ecological Modelling, 221(19), 2287-2301.
Hazen, E. L., Scales, K.L., Maxwell, S.M., Briscoe, D.K., Welch, H., Bograd, S.J., Bailey, H., Benson, S.R., Eguchi, T., Dewar, H., Kohin, S., Costa, D.P., Crowder, L.B., Lewison R.L. (2018). A dynamic ocean management tool to reduce bycatch and support sustainable fisheries.Science Advances, 4(5), DOI:10.1126/sciadv.aar3001
Hernandez, F., Bertino, L., Brassington, G., Chassignet, E., Cummings, J., Davidson, F., Drevillon, M., Garric, G., Kamachi, M., Lellouche, J.M., Mahdon, R., Martin, M.J,, Ratsimandresy, A., and Regnier, C. (2009). Validation and Inter-comparison studies within GODAE. Oceanography, 22(3): 128-143, https://doi.org/10.5670/oceanog.2009.71
Hernandez, F., Smith, G., Baetens, K., Cossarini, G., Garcia-Hermosa, I., Drevillon, M., von Schuckman, K. (2018). Measuring performances, skill and accuracy in operational oceanography: New challenges and approaches. In: "New Frontiers in Operational Oceanography", E. Chassignet, A. Pascual, J. Tintoré, and J. Verron, Eds., GODAE OceanView, 759-796, DOI:10.17125/gov2018.ch29
Hernandez, O., Lehodey, P., Senina, I., Echevin, V., Ayon, P., Bertrand, A., Gaspar, P. (2014). Understanding mechanisms that control fish spawning and larval recruitment: Parameter optimization of an Eulerian model (SEAPODYM-SP) with Peruvian anchovy and sardine eggs and larvae data. Progress in Oceanography, 123, 105-122, http://dx.doi.org/10.1016/j.pocean.2014.03.001
Hipsey, M.R., Gal, G., Arhonditsis, G.B., Carey, C.C., Elliott, J.A., Frassl, M. A., ... & Robson, B.J. (2020). A system of metrics for the assessment and improvement of aquatic ecosystem models. Environmental Modelling & Software, 128, 104697, https://doi.org/10.1016/j.envsoft.2020.104697
Hobday, A.J., Hartog, J.R., Timmiss, T., Fielding, J. (2010). Dynamic spatial zoning to manage southern bluefin tuna (Thunnus maccoyii) capture in a multi-species longline fishery. Fisheries Oceanography, 19(3), 243-253, https://doi.org/10.1111/j.1365-2419.2010.00540.x
Houtekamer, P. L., and Mitchell, H. L. (1998). Data Assimilation Using an Ensemble Kalman Filter Technique. Monthly Weather Review,126(3), 796-811, https://doi.org/10.1175/1520-0493(1998)126<0796:DAUAEK>2.0.CO;2
Howell, E.A., Kobayashi, D.R., Parker, D.M., Balazs, G.H., Polovina, J.J. (2008). TurtleWatch: a tool to aid in the bycatch reduction of loggerhead turtles Caretta caretta in the Hawaii-based pelagic longline fishery. Endangered Species Research, 5:267-278, https://doi.org/10.3354/esr00096
Hu, J., Fennel, K., Mattern, J.P. and Wilkin, J. (2012). Data assimilation with a local Ensemble Kalman Filter applied to a three-dimensional biological model of the Middle Atlantic Bight. Journal of Marine Systems, 94, 145-156, https://doi.org/10.1016/j.jmarsys.2011.11.016
Huse, G., Melle, W., Skogen, M.D., Hjøllo, S.S., Svendsen, E., and Budgell, W.P. (2018). Modeling Emergent Life Histories of Copepods. Frontiers in Ecology and Evolution, 6(23), https://doi.org/10.3389/fevo.2018.00023
IOCCG. (2020). Synergy between Ocean Colour and Biogeochemical/Ecosystem Models. Dutkiewicz, S. (ed.), IOCCG Report Series, No. 19, International Ocean Colour Coordinating Group, Dartmouth, Canada, http://dx.doi.org/10.25607/OBP-711
Ishizaka, J. (1990). Coupling of coastal zone color scanner data to a physical-biological model of the southeastern US continental shelf ecosystem: 2. An Eulerian model. Journal of Geophysical Research: Oceans, 95(C11), 20183-20199, https://doi.org/10.1029/JC095iC11p20183
Jaccard,P.,Hjermann,D.Ø.,Ruohola,J.,Marty, S.,Kristiansen, T., Sørensen,K.,Kaitala, S.,Mangin,A.,Pouliquen, S., et al. (2021). Quality Information Document For Global Ocean Reprocessed in-situ Observations of Biogeochemical Products INSITU_GLO_BGC_REP_OBSERVATIONS_013_046, http://doi.org/10.13155/54846, https://catalogue.marine.copernicus.eu/documents/QUID/CMEMS-INS-QUID-013-046.pdf
Jech, J.M., Horne, J.K., Chu, D., Demer, D.A., Francis, D.T.I., Gorska, N., Jones, B., Lavery, A.C., Stanton, T.K., Macaulay, G.J., Reeder, D.B., Sawada, K. (2015). Comparisons among ten models of acoustic backscattering used in aquatic ecosystems. The Journal of the Acoustical Society of America, 138: 3742, https://doi.org/10.1121/1.4937607
Jennings, S., and Collingridge, K. (2015). Predicting Consumer Biomass, Size-Structure, Production, Catch Potential, Responses to Fishing and Associated Uncertainties in the World’s Marine Ecosystems. PLoS ONE, 10(7), e0133794. https://doi.org/10.1371/journal.pone.0133794
Jolliff, J.K., Kindle, J.C., Shulman, I., Penta, B., Friedrichs, M.A.M., Helber, R.W., and Arnone, R. (2009). Summary diagrams for coupled hydrodynamic-ecosystem model skill assessment. Journal of Marine Systems, 76, 64-82, https://doi.org/10.1016/j.jmarsys.2008.05.014
Jones, E.M., Baird, M.E., Mongin, M., Parslow, J., Skerratt, J., Lovell, J., Margvelashvili, N., Matear, R.J., Wild-Allen, K., Robson, B. and Rizwi, F. (2016). Use of remote-sensing reflectance to constrain a data assimilating marine biogeochemical model of the Great Barrier Reef. Biogeosciences, 13(23), 6441, https://doi.org/10.5194/bg-13-6441-2016
Kaufman, D.E. (2017). Using High-Resolution Glider Data and Biogeochemical Modeling to Investigate Phytoplankton Variability in the Ross Sea. Dissertations, Theses, and Masters Projects, William & Mary, Paper 1499449869, http://dx.doi.org/10.21220/M2BK8V
Kriest, I., Kähler, P., Koeve, W., Kvale, K., Sauerland, V., and Oschlies, A. (2020). One size fits all? Calibrating an ocean biogeochemistry model for different circulations. Biogeosciences, 17, 3057-3082, https://doi.org/10.5194/bg-17-3057-2020
Krishnamurthy, A., Moore, J. K., Mahowald, N., Luo, C., and Zender, C.S. (2010). Impacts of atmospheric nutrient inputs on marine biogeochemistry. Journal of Geophysical Research: Biogeosciences, 115(G1), https://doi.org/10.1029/2009JG001115
Lambert, C., Mannocci, L., Lehodey, P., Ridoux, V. (2014). Predicting Cetacean Habitats from Their Energetic Needs and the distribution of Their Prey in Two Contrasted Tropical Regions. PLoS ONE, 9(8), e105958, https://doi.org/10.1371/journal.pone.0105958
Lamouroux, J., Perruche, C., Mignot, A., Paul, J., Szczypta, C. (2019). Quality Information Document For Global Biogeochemical Analysis and Forecast Product, https://catalogue.marine.copernicus.eu/documents/QUID/CMEMS-GLO-QUID-001-028.pdf
Lamouroux, J., Perruche, C., Mignot, A., Gutknecht, E., Ruggiero, G., Evaluation of the CMEMS global biogeochemical simulation, with assimilation of satellite Chla concentrations, in prep.
Laws, E.A. (2013). Evaluation of In Situ Phytoplankton Growth Rates: A Synthesis of Data from Varied Approaches. Annual Review of Marine Science, 5(1), 247-268, https://doi.org/10.1146/annurev-marine-121211-172258
Le Quéré, C., Harrison, S. P., Prentice, I. C., Buitenhuis, E. T., Aumont, O., Bopp, L., Claustre, H., Cotrim Da Cunha, L., Geider, R., Giraud, X., Klaas, C., Kohfeld, K. E., Legendre, L., Manizza, M., Platt, T., Rivkin, R. B., Sathyendranath, S., Uitz, J., Watson, A. J., and Wolf‐Gladrow, D. (2005). Ecosystem dynamics based on plankton functional types for global ocean biogeochemistry models. Global Change Biology, 11, 2016-2040, https://doi.org/10.1111/j.1365-2486.2005.1004.x
Le Traon et al. (2017). The Copernicus marine environmental monitoring service: main scientific achievements and future prospects. Special Issue Mercator Océan International #56. Available at: https://marine.copernicus.eu/news/mercator-ocean-journal-56-cmems-special-issue
Le Traon, P. Y., Reppucci, A., Fanjul, E. A., Aouf, L., Behrens, A., Belmonte, M., Bentamy, A., Bertino, L., Brando, V. E., Kreiner, M. B., Benkiran, M., Carval, T., Ciliberti, S. A., Claustre, H., Clementi, E., Coppini, G., Cossarini, G., De Alfonso Alonso- Muñoyerro, M., Delamarche, A., Dibarboure, G., Dinessen, F., Drevillon, M., Drillet, Y., Faugere, Y., Fernández, V., Fleming, A., Garcia-Hermosa, M. I., Sotillo, M. G., Garric, G., Gasparin, F., Giordan, C., Gehlen, M., Grégoire, M., Guinehut, S., Hamon, M., Harris, C., Hernandez, F., Hinkler, J. B., Hoyer, J., Karvonen, J., Kay, S., King, R., Lavergne, T., Lemieux-Dudon, B., Lima, L., Mao, C., Martin, M. J., Masina, S., Melet, A., Nardelli, B. B., Nolan, G., Pascual, A., Pistoia, J., Palazov, A., Piolle, J. F., Pujol, M. I., Pequignet, A. C., Peneva, E., Gómez, B. P., de la Villeon, L. P., Pinardi, N., Pisano, A., Pouliquen, S., Reid, R., Remy, E., Santoleri, R., Siddorn, J., She, J., Staneva, J., Stoffelen, A., Tonani, M., Vandenbulcke, L., von Schuckmann, K., Volpe, G., Wettre, C., and Zacharioudaki, A. (2019). From observation to information and users: The Copernicus Marine Service Perspective. Frontiers in Marine Science, https://doi.org/10.3389/fmars.2019.00234
Lehodey, P., Senina, I., Murtugudde, R. (2008). A Spatial Ecosystem And Populations Dynamics Model (SEAPODYM) - Modelling of tuna and tuna-like populations. Progress in Oceanography, 78, 304-318, https://doi.org/10.1016/j.pocean.2008.06.004
Lehodey, P., Senina, I., Wibawa, T.A., Titaud, O., Calmettes, B., Tranchant, B., and Gaspar, P. (2017). Operational modelling of bigeye tuna (Thunnus obesus) spatial dynamics in the Indonesian region. Marine Pollution Bulletin, 131, 19-32, https://doi.org/10.1016/j.marpolbul.2017.08.020
Lehodey, P., Conchon, A., Senina, I., Domokos, R., Calmettes, B., Jouanno, J., Hernandez, O., and Kloser, R. (2015). Optimization of a micronekton model with acoustic data. ICES Journal of Marine Science, 72(5), 1399-1412, https://doi.org/10.1093/icesjms/fsu233
Lehodey, P., Murtugudde, R., and Senina, I. (2010). Bridging the gap from ocean models to population dynamics of large marine predators: A model of mid-trophic functional groups. Progress in Oceanography, 84, 69-84.
Lehodey, P., Senina, I., Calmettes, B, Hampton, J, Nicol S. (2013). Modelling the impact of climate change on Pacific skipjack tuna population and fisheries. Climatic Change, 119 (1): 95-109, DOI 10.1007/s10584-012-0595-1
Lengaigne, M., Menkes, C., Aumont, O., Gorgues, T., Bopp, L., André, J. M., and Madec, G. (2007). Influence of the oceanic biology on the tropical Pacific climate in a coupled general circulation model. Climate Dynamics, 28(5), 503-516. https://doi.org/10.1007/s00382-006-0200-2
Libralato, S., and Solidoro, C. (2009). Bridging biogeochemical and food web models for an Endto-End representation of marine ecosystem dynamics: The Venice lagoon case study. Ecological Modelling, 220(21), 2960-2971, https://doi.org/10.1016/j.ecolmodel.2009.08.017
Longhurst, A. (1998). Ecological geography in the sea. Academic Press.
Mattern, J. P., Fennel, K., and Dowd, M. (2012). Estimating time-dependent parameters for a biological ocean model using an emulator approach. Journal of Marine Systems, 96, 32-47, https://doi.org/10.1016/j.jmarsys.2012.01.015
Mattern, J.P., Dowd, M. and Fennel, K. (2013). Particle filter-based data assimilation for a three-dimensional biological ocean model and satellite observations. Journal of Geophysical Research: Oceans, 118(5), 2746-2760, https://doi.org/10.1002/jgrc.20213
Maunder, M.N., Punt, A.E. (2013). A review of integrated analysis in fisheries stock assessment. Fisheries Research, 142, 61-74, https://doi.org/10.1016/j.fishres.2012.07.025
Maury, O., Faugeras, B., Shin, Y.-J., Poggiale, J.-C., Ben Ari, T., and Marsac, F. (2007). Modeling environmental effects on the size-structured energy flow through marine ecosystems. Part 1: The model. Progress in Oceanography, 74(4), 479-499, https://doi.org/10.1016/j.pocean.2007.05.002
McEwan, R., S. Kay, D. Ford (2021). Quality Information Document For the Atlantic - European North West Shelf Production Centre, https://catalogue.marine.copernicus.eu/documents/QUID/CMEMS-NWS-QUID-004-002.pdf
Melsom, A. and Ç. Yumruktepe (2021), Quality Information Document For the Arctic Production Centre, https://catalogue.marine.copernicus.eu/documents/QUID/CMEMS-ARC-QUID-002-004.pdf, https://doi.org/10.48670/moi-00003
Mignot, A., Claustre, H., Cossarini, G., D’Ortenzio, F., Gutknecht, E., Lamouroux, J., Lazzari, P., Perruche, C., Salon, S., Sauzède, R., Taillandier, V., Teruzzi, A. (2021). Defining BGC-Argo-based metrics of ocean health and biogeochemical functioning for the evaluation of global ocean models. Biogeosciences, https://doi.org/10.5194/bg-2021-2
Miller, C.B., Lynch, D.R., Carlotti, F., Gentleman, W., Lewis, C.V.W. (1998). Coupling of an individual-based population dynamic model of Calanus finmarchicus to a circulation model for theGeorges Bank region. Fisheries Oceanography, 7(3-4), 219-234, https://doi.org/10.1046/j.1365-2419.1998.00072.x
Mogensen, K.S., Balmaseda, M.A., Weaver, A., Martin, M.J., Vidard, A. (2009). NEMOVAR: A variational data assimilation system for the NEMO ocean model. ECMWF Newsl., 120, 17-21.
Moore, A. M., Martin, M. J., Akella, S., Arango, H. G., Balmaseda, M., Bertino, L., ... and Weaver, A. T. (2019). Synthesis of ocean observations using data assimilation for operational, real time and reanalysis systems: A more complete picture of the state of the ocean. Frontiers in Marine Science, 6:90, https://doi.org/10.3389/fmars.2019.00090
Moriarty, R., and O’Brien, T. (2013). Distribution of mesozooplankton biomass in the global ocean. Earth System Science Data, 5(1), 45-55, https://doi.org/10.5194/essd-5-45-2013
Natvik, L., and Evensen, G. (2003). Assimilation of ocean colour data into a biochemical model of the North Atlantic. Part 1: Data assimilation experiments. Journal of Marine Systems, 41, 127-153, https://doi.org/10.1016/S0924-7963(03)00016-2
Nerger, L. andGregg,W.W.(2007).AssimilationofSeaWiFSdata intoa globalocean-biogeochemicalmodelusing a local SEIK filter. Journal of Marine Systems, 68, 237-254, https://doi.org/10.1016/j.jmarsys.2006.11.009
Nerger, L., and Gregg, W.W. (2008). Improving assimilation of SeaWiFS data by the application of bias correction with a local SEIK filter. Journal of Marine Systems, 73, 87-102, https://doi.org/10.1016/j.jmarsys.2007.09.007
Nerger, L., Janjić, T., Schröter, J., and Hiller, W. (2012). A Unification of Ensemble Square Root Kalman Filters. Monthly Weather Review, 140(7), 2335-2345, https://doi.org/10.1175/MWR-D-11-00102.1
Neumann, T. (2000). Towards a 3D-ecosystem model of the Baltic Sea. Journal of Marine Systems, 25(3), 405-419, https://doi.org/10.1016/S0924-7963(00)00030-0
Olsen, A., Lange, N., Key, R. M., Tanhua, T., Bittig, H. C., Kozyr, A., Àlvarez, M., Azetsu-Scott, K., Becker, S., Brown, P. J., Carter, B. R., Cotrim da Cunha, L., Feely, R. A., van Heuven, S., Hoppema, M., Ishii, M., Jeansson, E., Jutterström, S., Landa, C. S., Lauvset, S. K., Michaelis, P., Murata, A., Pérez, F. F., Pfeil, B., Schirnick, C., Steinfeldt, R., Suzuki, T., Tilbrook, B., Velo, A., Wanninkhof, R. and Woosley, R. J. (2020). GLODAPv2.2020 - the second update of GLODAPv2. Earth System Science Data, https://doi.org/10.5194/essd-12-3653-2020
Ourmières, Y., Brasseur, P., Lévy, M., Brankart, J.M. and Verron, J. (2009). On the key role of nutrient data to constrain a coupled physical-biogeochemical assimilative model of the North Atlantic Ocean. Journal of Marine Systems, 75(1-2), 100-115, https://doi.org/10.1016/j.jmarsys.2008.08.003
Palmer, J. R., and Totterdell, I. J. (2001). Production and export in a global ocean ecosystem model. Deep Sea Research Part I: Oceanographic Research Papers, 48(5), 1169-1198, https://doi.org/10.1016/S0967-0637(00)00080-7
Park, J.-Y., Stock, C. A., Yang, X., Dunne, J. P., Rosati, A., John, J., et al. (2018). Modeling global ocean biogeochemistry with physical data assimilation: A pragmatic solution to the equatorial instability. Journal of Advances in Modeling Earth Systems, 10, 891- 906, https://doi.org/10.1002/2017MS001223
Pérez-Jorge, S., Tobeña, M., Prieto, R., Vandeperre, F., Calmettes, B., Lehodey, P., Silva, M.A. (2020). Environmental drivers of large-scale movements of baleen whales in the mid-North Atlantic Ocean. Diversity and Distributions, 26(6), 683-698, https://doi.org/10.1111/ddi.13038
Petrik, C.M., Stock, C.A., Andersen, K.H., van Denderen, P.D., and Watson, J.R. (2019). Bottom-up drivers of global patterns of demersal, forage, and pelagic fishes. Progress in Oceanography, 176:102124, https://doi.org/10.1016/j.pocean.2019.102124
Petrik, C.M., Stock, C.A., Andersen, K.H., van Denderen, P.D., and Watson, J.R. (2020). Large pelagic fish are most sensitive to climate change despite pelagification of ocean foodwebs. Frontiers in Marine Science, 7, 588482, https://doi.org/10.3389/fmars.2020.588482
Pham, D.T., Verron, J., Roubaud, M.C. (1998). A singular evolutive extended Kalman filter for data assimilation in oceanography. Journal of Marine Systems, 16(3-4): 323-340, https://doi.org/10.1016/S0924-7963(97)00109-7
Pradhan, H.K., Völker, C., Losa, S.N., Bracher, A., and Nerger, L. (2020). Global assimilation of oceancolor data of phytoplankton functional types: Impact of different data sets. Journal of Geophysical Research: Oceans, 125, e2019JC015586, https://doi.org/10.1029/2019JC015586
Proud, R., Handegard, N. O., Kloser, R. J., Cox, M. J., and Brierley, A. S. (2018). From siphonophores to deep scattering layers:Uncertainty ranges for the estimation of global mesopelagic fish biomass. ICES Journal of Marine Science, 76, 718-733, https://doi.org/10.1093/icesjms/fsy037
Redfield, A.C. (1934). On the proportions of organic derivatives in sea water and their relation to the composition of plankton. James Johnson Memorial Volume, R. J. Daniel, Ed., University Press of Liverpool, 177-192.
Roberts, J. J., Best, B. D., Mannocci, L., Fujioka, E., Halpin, P. N., Palka, D. L., … Lockhart, G. G. (2016). Habitat-based cetacean density models for the U.S. Atlantic and Gulf of Mexico. Nature Scientific Reports, 6, 22615, https://doi.org/10.1038/srep22615
Romagosa M., Lucas C., Pérez-Jorge S., Tobeña M., Lehodey P., Reis J., Cascão I., Lammers M. O., Caldeira R. M. A., Silva M. A. (2020). Differences in regional oceanography and prey biomass influence the presence of foraging odontocetes at two Atlantic seamounts. Marine Mammal Science, 36: 158-179, https://doi.org/10.1111/mms.12626
Romagosa, M., Pérez-Jorge, S., Cascão, I., Mouriño, H., Lehodey, P., Marques, T.A, Silva, M.A. (2021). Food talk: 40-Hz fin whale calls are associated with prey availability. Proceedings of the Royal Society B Biological Sciences, 288(1954), https://doi.org/10.1098/rspb.2021.1156
Rose, K.A., Fiechter, J., Curchitser, E.N., Hedstrom, K., Bernal, M., Creekmore, S., Haynie, A.C., Ito, S., Lluch-Cota, S.E., Megrey, B.A., Edwards, C.A., Checkley, D.M., Koslow, T., Mcclatchie, S., Werner, F.E., Maccall, A.D., and Agostini, V.N. (2015). Demonstration of a fully-coupled end-to-end model for small pelagic fish using sardine and anchovy in the California Current. Progress in Oceanography, 138, 348-380, https://doi.org/10.1016/j.pocean.2015.01.012
Russell, J., Sarmiento, J., Cullen, H., Hotinski, R., Johnson, K.S., Riser, S.C., et al. (2014). The Southern Ocean Carbon and Climate Observations and Modeling Program (SOCCOM). Ocean Carbon Biogeochem. News. Available at https://web.whoi.edu/ocb/wp-content/uploads/sites/43/2016/12/OCB_NEWS_FALL14.pdf
Ryabinin, V., Barbière, J., Haugan, P., Kullenberg, G., Smith, N., McLean, C., Troisi, A., Fischer, A., Aricò, S., Aarup, T., Pissierssens, P., Visbeck, M., Enevoldsen, H.O., Rigaud, J. (2019). The UN Decade of Ocean Science for Sustainable Development, Frontiers in Marine Science, 6,470, https://doi.org/10.3389/fmars.2019.00470
Sakov, P., and Oke, P.R. (2008). A deterministic formulation of the ensemble Kalman filter: an alternative to ensemble square root filters. Tellus A, 60, 361-371, https://doi.org/10.1111/j.1600-0870.2007.00299.x
Salon, S., Cossarini, G., Bolzon, G., Feudale, L., Lazzari, P., Teruzzi, A., Solidoro, C., Crise, A. (2019). Novel metrics based on Biogeochemical Argo data to improve the model uncertainty evaluation of the CMEMS Mediterranean marine ecosystem forecasts. Ocean Science, 15, 997-1022, https://doi.org/10.5194/os-15-997-2019
Santana-Falcon, Y., Brasseur, P., Brankart, J.M., and Garnier, F. (2020). Assimilation of chlorophyll data into a stochastic ensemble simulation for the North Atlantic ocean, Ocean Science, 16, 1297-1315, https://doi.org/10.5194/os-16-1297-2020
Schartau, M., Wallhead, P., Hemmings, J., Löptien, U., Kriest, I., Krishna, S., Ward, B. A., Slawig, T., and Oschlies, A. (2017). Reviews and syntheses: parameter identification in marine planktonic ecosystem modelling. Biogeosciences, 14, 1647-1701, https://doi.org/10.5194/bg-14-1647-2017
Scutt Phillips, J., Sen Gupta, A., Senina, I., van Sebille, E., Lange, M., Lehodey, P., Hampton, J., Nicol, S. (2018). An individual-based model of skipjack tuna (Katsuwonus pelamis) movement in the tropical Pacific Ocean. Progress in Oceanography, 164, 63-74, https://doi.org/10.1016/j.pocean.2018.04.007
Senina, I., Lehodey, P., Hampton, J., Sibert, J. (2019). Quantitative modelling of the spatial dynamics of South Pacific and Atlantic albacore tuna populations. Deep Sea Research Part II: Topical Studies in Oceanography, 175, 104667, https://doi.org/10.1016/j.dsr2.2019.104667
Senina, I., Lehodey, P., Sibert, J., Hampton, J. (2020). Improving predictions of a spatially explicit fish population dynamics model using tagging data. Canadian Journal of Aquatic and Fisheries Sciences, 77(3), 576-593, https://doi.org/10.1139/cjfas-2018-0470
Senina, I., Sibert, J., and Lehodey, P. (2008). Parameter estimation for basin-scale ecosystem-linked population models of large pelagic predators: Application to skipjack tuna. Progress in Oceanography, 78, 319-335, https://doi.org/10.1016/j.pocean.2008.06.003
Sibert, J., Senina, I., Lehodey, P., Hampton, J. (2012). Shifting from marine reserves to maritime zoning for conservation of Pacific bigeye tuna (Thunnus obesus). Proceedings of the National Academy of Sciences, 109(44): 18221-18225.
Simon, E. and Bertino, L. (2009). Application of the Gaussian anamorphosis to assimilation in a 3-D coupled physical-ecosystem model of the North Atlantic with the EnKF: a twin experiment. Ocean Science, 5(4), 495-510, https://doi.org/10.5194/os-5-495-2009
Simon, E. Bertino, L. (2012). Gaussian anamorphosis extension of the DEnKF for combined state parameter estimation: Application to a 1D ocean ecosystem model. Journal of Marine Systems, 89(1), 1-18, https://doi.org/10.1016/j.jmarsys.2011.07.007
Simon, E., Samuelsen, A., Bertino, L. and Mouysset, S. (2015). Experiences in multiyear combined state-parameter estimation with an ecosystem model of the North Atlantic and Arctic Oceans using the Ensemble Kalman Filter. Journal of Marine Systems, 152, 1–17, https://doi.org/10.1016/j.jmarsys.2015.07.004
Skákala, J., Ford, D., Brewin, R. J. W., McEwan, R., Kay, S., Taylor, B., Mora, L., and Ciavatta, S. (2018). The assimilation of phytoplankton functional types for operational forecasting in the northwest European shelf. Journal of Geophysical Research: Oceans, 123, 5230-5247, https://doi.org/10.1029/2018JC014153
Skakala, J., Bruggeman, J., Brewin, R.J., Ford, D.A. and Ciavatta, S. (2020). Improved representation of underwater light field and its impact on ecosystem dynamics: A study in the North Sea. Journal of Geophysical Research: Oceans, 125(7), e2020JC016122, https://doi.org/10.1029/2020JC016122
Skákala, J., Ford, D., Bruggeman, J., Hull, T., Kaiser, J., King, R.R., Loveday, B., Palmer, M.R., Smyth, T., Williams, C.A. and Ciavatta, S. (2021a). Towards a multi-platform assimilative system for North Sea biogeochemistry. Journal of Geophysical Research: Oceans, 126(4), e2020JC016649, https://doi.org/10.1029/2020JC016649
Skakala, J., Bruggeman, J., Ford, D.A., Wakelin, S.L., Akpınar, A., Hull, T., Kaiser, J., Loveday, B.R., Williams, C.A.J. and Ciavatta, S. (2021b). Improved consistency between the modelling of ocean optics, biogeochemistry and physics, and its impact on the North-West European Shelf seas. Earth andSpace Science Open Archive ESSOAr, https://doi.org/10.1002/essoar.10506737.2
Skogen, M.D. (1993). A User's guide to NORWECOM (the NORWegian ECOlogical Model system). Technical report 6, Inst.of Marine Research, Division of Marine Env., Pb 1870, N-5024 Bergen, Norway.
Skogen, M.D. and Søiland, H. (1998). A User's guide to NORWECOM v2.0 (the NORWegian ECOlogical Model system). Tech.rep. Fisken og Havet 18, Inst. of Marine Research, Pb.1870, N-5024 Bergen. Norway.
Song, H., Edwards, C.A., Moore, A.M., and Fiechter, J. (2016). Data assimilation in a coupled physical-biogeochemical model of the California current system using an incremental lognormal 4-dimensional variational approach: part 3 - Assimilation in a realistic context using satellite and in situ observations. Ocean Modelling, 106, 1531-145, https://doi.org/10.1016/j.ocemod.2016.04.001
Spindler, M. (1994). Notes on the biology of sea ice in the Arctic and Antarctic. Polar Biology, 14, 319-324, https://doi.org/10.1007/BF00238447
Storto, A., Oddo, P., Cipollone, A., Mirouze, I. and Lemieux-Dudon, B. (2018). Extending an oceanographic variational scheme to allow for affordable hybrid and four-dimensional data assimilation. Ocean Modelling, 128,.67-86, https://doi.org/10.1016/j.ocemod.2018.06.005
Stow, C. A., Jolliff, J., McGillicuddy Jr, D. J., Doney, S. C., Allen, J. I., Friedrichs, M. A., Wallhead, P., 2009. Skill assessment for coupled biological/physical models of marine systems. Journal of Marine Systems, 76(1-2), 4-15, https://doi.org/10.1016/j.jmarsys.2008.03.011
Taylor, K.E. (2001). Summarizing multiple aspects of model performance in a single diagram. Journal of Geophysical Research: Atmospheres, 106(D7), 7183-7192, https://doi.org/10.1029/2000JD900719
Teruzzi, A, Di Cerbo, P, Cossarini, G, Pascolo, E, Salon, S. (2019). Parallel implementation of a data assimilation scheme for operational oceanography: The case of the MedBFM model system. Computers & Geosciences, 124, 103-114, https://doi.org/10.1016/j.cageo.2019.01.003
Teruzzi, A., Dobricic, S., Solidoro, C., and Cossarini, G. (2014). A 3-D variational assimilation scheme in coupled transport-biogeochemical models: Forecast of Mediterranean biogeochemical properties. Journal of Geophysical Research: Oceans, 119, 200-217, https://doi.org/10.1002/2013JC009277
Tissier, A.-S., Brankart, J.M., Testut, C.E., Ruggiero, G., Cosme, E., and Brasseur, P. (2019). A multiscale ocean data assimilation approach combining spatial and spectral localization. Ocean Science, 15, 443-457, https://doi.org/10.5194/os-15-443-2019
Torres, R., Allen, J.I. and Figueiras, F.G. (2006). Sequential data assimilation in an upwelling influenced estuary. Journal of Marine Systems, 60(3-4), 317-329, https://doi.org/10.1016/j.jmarsys.2006.02.001
Uitz, J., Claustre, H., Morel, A., and Hooker, S. B. (2006). Vertical distribution of phytoplankton communities in open ocean: An assessment based on surface chlorophyll. Journal of Geophysical Research: Oceans, 111(C8), https://doi.org/10.1029/2005JC003207
van Leeuwen, P.J. (2010). Nonlinear data assimilation in geosciences: an extremely efficient particle filter. Quarterly Journal of the Royal Meteorological Society, 136(653), 1991-1999, https://doi.org/10.1002/qj.699
Vandenbulcke, L., and Barth, A. (2015). A stochastic operational system of the Black Sea: Tech. and validation. Ocean Modelling, 93, 7-21.
Verdy, A., and Mazloff, M.R. (2017). A data assimilating model for estimating Southern Ocean biogeochemistry. Journal of Geophysical Research: Oceans, 122(9), 6968-6988, https://doi.org/10.1016/j.ocemod.2015.07.010
Vetra-Carvalho, S., Van Leeuwen, P.J., Nerger, L., Barth, A., Altaf, M.U., Brasseur, P., Kirchgessner, P. and Beckers, J.M. (2018). State-of-the-art stochastic data assimilation methods for high-dimensional non-Gaussian problems. Tellus A: Dynamic Meteorology and Oceanography, 70(1), 1-43, https://doi.org/10.1080/16000870.2018.1445364
Vichi, M., Cossarini, G., Gutierrez Mlot, E., Lazzari, P., Lovato, T., Mattia, G., Masina, S., McKiver, W., Pinardi, N., Solidoro, C., Zavatarelli, M. (2015). The Biogeochemical Flux Model (BFM): Equation Description and User Manual. BFM version 5.1. BFM Report series N.1, March 2015, Bologna, Italy, pp. 89.
Wang, B., Fennel, K., Yu, L. and Gordon, C. (2020). Assessing the value of biogeochemical Argo profiles versus ocean color observations for biogeochemical model optimization in the Gulf of Mexico. Biogeosciences, 17(15), 4059-4074, https://doi.org/10.5194/bg-17-4059-2020
Warner, J.C., Armstrong, B., He, R., Zambon, J.B. (2010). Development of a Coupled Ocean–Atmosphere–Wave–Sediment Transport (COAWST) Modeling System. Ocean Modelling, 35(3), 230-244, https://doi.org/10.1016/j.ocemod.2010.07.010
Waters, J., Lea, D.J., Martin, M.J., Mirouze, I., Weaver, A. and While, J. (2015). Implementing a variational data assimilation system in an operational 1/4 degree global ocean model. Quarterly Journal of the Royal Meteorological Society, 141(687), 333-349, https://doi.org/10.1002/qj.2388
Waters, J., Bell, M. J., Martin, M. J., and Lea, D. J. (2017). Reducing ocean model imbalances in the equatorial region caused by data assimilation. Quarterly Journal of the Royal Meteorological Society, 143(702), 195-208.
While, J., Totterdell, I. and Martin, M. (2012). Assimilation of pCO2 data into a global coupled physical-biogeochemical ocean model. Journal of Geophysical Research: Oceans, 117(C3), https://doi.org/10.1029/2010JC006815
Wright, R.M., Le Quéré, C., Buitenhui, E., Pitois, S., Gibbons, M. (2021). Role of jellyfish in the plankton ecosystem revealed using a global ocean biogeochemical model. Biogeosciences, 18, 1291-1320, https://doi.org/10.5194/bg-18-1291-2021
Xiao, Y., and M.A.M. Friedrichs (2014). Using biogeochemical data assimilation to assess the relative skill of multiple ecosystem models: effects of increasing the complexity of the planktonic food web. Biogeosciences, 11(11), 3015-3030, https://doi.org/10.5194/bg-11-3015-2014
Yool, A., Popova, E. E., and Anderson, T. R. (2013). MEDUSA-2.0: an intermediate complexity biogeochemical model of the marine carbon cycle for climate change and ocean acidification studies. Geoscientific Model Development, 6, 1767-1811, https://doi.org/10.5194/gmd-6-1767-2013
Yu, L., Fennel, K., Bertino, L., El Gharamti, M., and Thompson, K. R. (2018). Insights on multivariate updates of physical and biogeochemical ocean variables using an Ensemble Kalman Filter and an idealized model of upwelling. Ocean Modelling, 126, 13-28, doi:10.1016/j.ocemod.2018.04.005
Zeebe, R., and Wolf-Gladrow, D. (2001). CO2 in Seawater: Equilibrium, Kinetics, Isotopes. Elsevier Oceanography Book Series, 65, 346 pp, Amsterdam.
Zhou M., Carlotti F., Zhu Y. (2010). A size-spectrum zooplankton closure model for ecosystem modelling. Journal of Plankton Research, 32(8), 1147-1165, https://doi.org/10.1093/plankt/fbq054


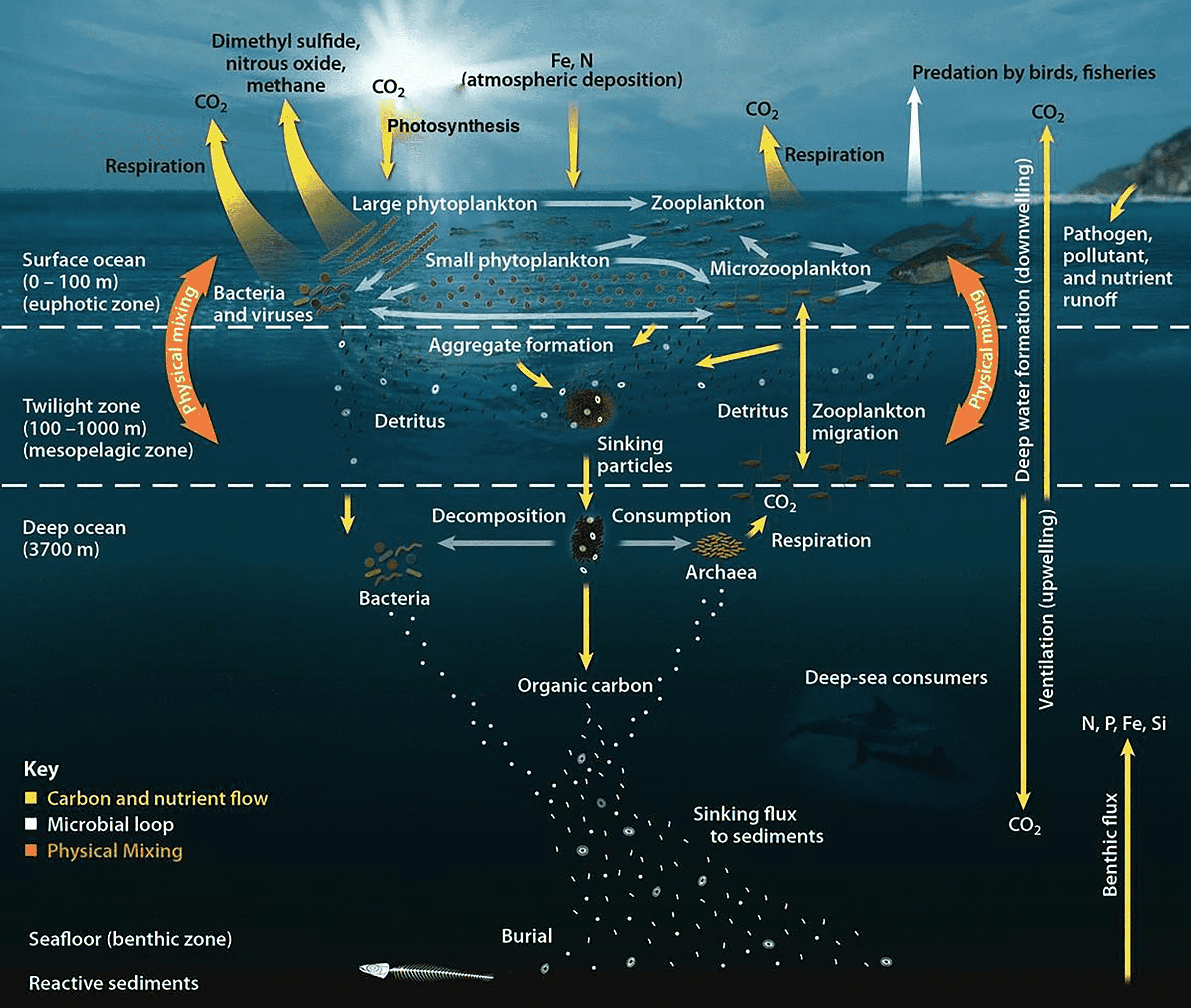
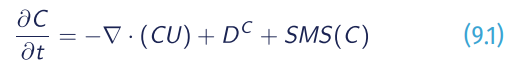
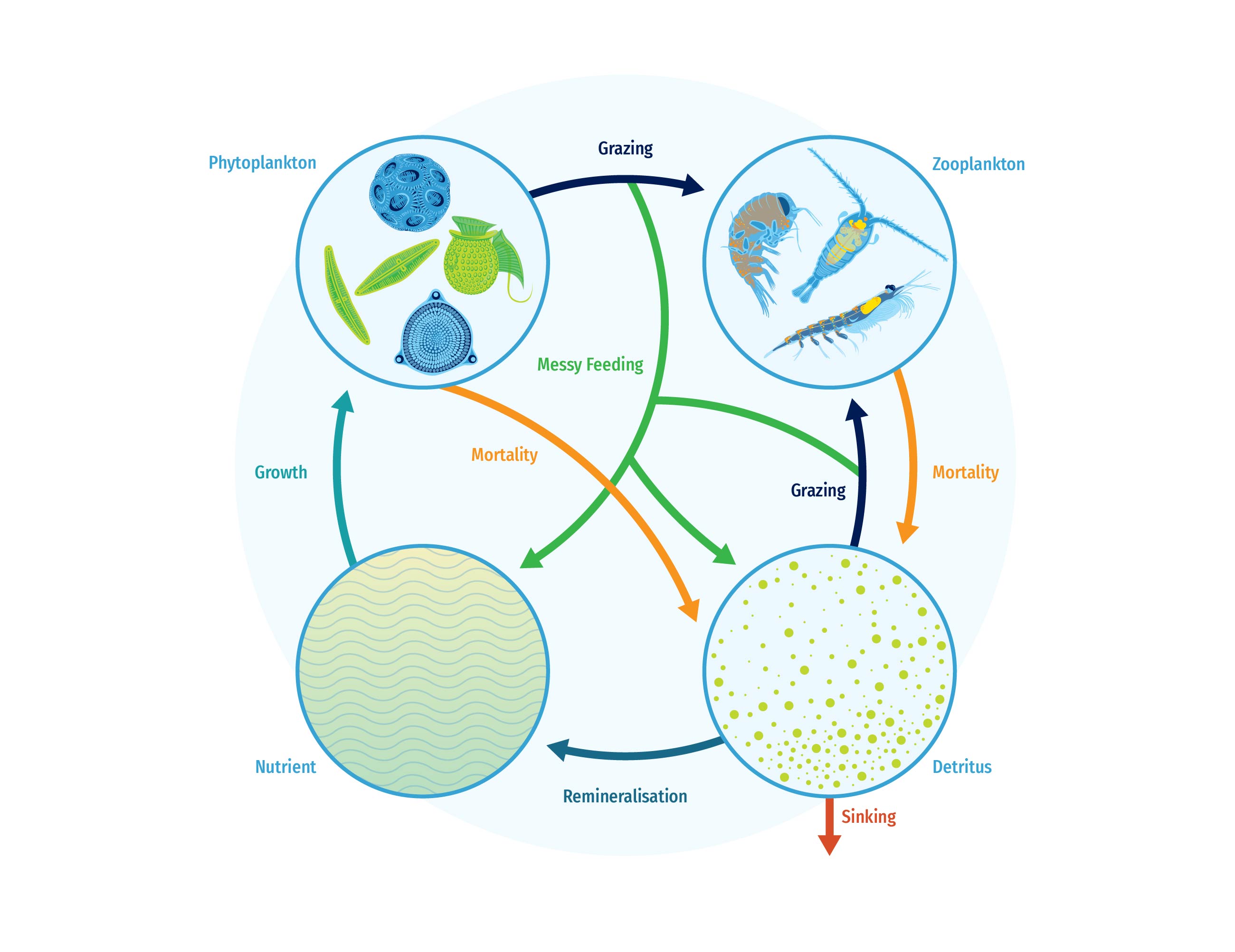


Follow us